PEROXISOME BIOGENESIS MODELS
The proliferation of peroxisomes is a striking feature of the methylotrophic yeast, Pichia pastoris, when cells are shifted from glucose medium (non-peroxisome-requiring media) to methanol or oleate medium (peroxisome-requiring media). Glucose-grown cells contain only 1-3 small peroxisomes, a single-membrane bound organelle that contains a wide range of metabolic enzymes involved in such processes as ß-oxidation of fatty acids (like oleate), elimination of hydrogen peroxide and oxidation of methanol. Upon shift from glucose to methanol or oleate, cells induce peroxisomes to cope with the task of surviving under these conditions: peroxisomes proliferate and import the necessary metabolic enzymes. The importance of this step is shown by the fact that mutations in either peroxins (proteins involved in the biogenesis of peroxisomes or transport of proteins into the peroxisomal matrix) or metabolic enzymes lead to severe growth defects on these peroxisome-requiring media (for review see Subramani, 1998).
Peroxisome morphology on different growth media (Pichia pastoris) |
Glucose
|
Methanol |
Oleate |
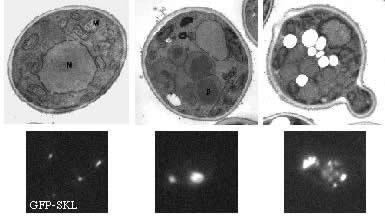
|
In humans, functional peroxisomes are also required for a wide range of metabolic pathways, like ß-oxidation of fatty acids, elimination of hydrogen peroxide, synthesis of plasmalogens and cholesterol. The importance of peroxisomes is underscored by the severe effects of peroxisomal dysfunction. This is most clearly demonstrated in the human genetic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum's disease which are characterized by mental retardation, severe neurologic, hepatic and renal abnormalities, and premature mortality (Lazarow and Moser, 1989).
Components of the peroxisomal matrix and membrane protein import machinery.
Like the sorting of proteins to other subcellular compartments, protein targeting to peroxisomes is signal dependent. The PTS1 and PTS2 signals direct proteins to the peroxisome matrix, whereas mPTSs specify a peroxisomal membrane location (Lazarow, 2003). These PTSs are recognized by soluble, cytosolic receptors – Pex5p for PTS1 (McCollum et al., 1993; Terlecky et al., 1995), Pex7p and its co-receptor, Pex20p, for PTS2 (Leon et al., 2006; Rehling et al., 1996; Stein et al., 2002; Titorenko et al., 1998; Zhang et al., 1996) and Pex19p (and/or other undefined components) for mPTSs (Jones et al., 2004; Snyder et al., 2000). Following cargo recognition, receptor/cargo complexes are delivered to the peroxisome membrane for further action.
The peroxisome membrane has many peroxins that facilitate the import of matrix and membrane proteins. Two subcomplexes, known as the docking (Pex8p, Pex13p, Pex14p, Pex17p) and RING (Pex2, Pex10p, Pex12p) subcomplexes, are bridged by Pex8p or another protein, Pex3p, to form a larger complex known as the importomer (Agne et al., 2003; Hazra et al., 2002). Pex3p also acts as the peroxisomal docking site for Pex19p (Fang et al., 2004).
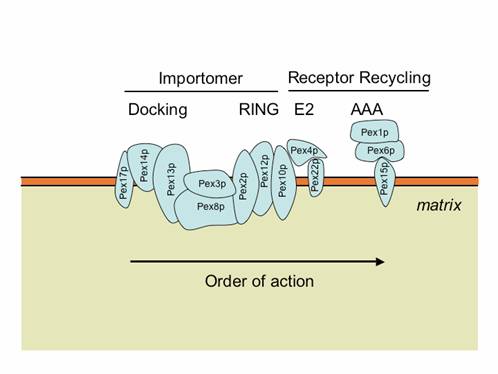
The importomer plays a role in matrix, but not membrane, protein import. The PTS1 and PTS2 receptors and their accessory proteins (e.g. Pex20p) ferry cargo from the cytosol and first interact with the docking subcomplex (Heiland and Erdmann, 2005). The receptor/cargo complexes then either enter the matrix, or are deeply embedded in the peroxisome membrane (Dammai and Subramani, 2001; Leon et al., 2006; Miyata and Fujiki, 2005; Nair et al., 2004). This is followed by cargo release into the peroxisome matrix, export/release of the receptors on the peroxisome membrane (Leon et al., 2006; Miyata and Fujiki, 2005; Platta et al., 2005), followed by dislocation/recycling of the receptors from a peroxisome-associated state to the cytosol (Leon et al., 2006; Miyata and Fujiki, 2005; Platta et al., 2005). Mutations in any component of the importomer affect the import of peroxisomal matrix proteins, suggesting that the whole importomer is somehow involved in protein translocation across this membrane (Agne et al., 2003). However, certain transient residents of the peroxisome matrix, such as Pex5p and Pex20p, become peroxisome-associated and protease protected, even in the absence of the RING subcomplex of the importomer, but their entry into the peroxisome is Pex14p-dependent (Leon et al., 2006; Zhang et al., 2006). These data suggest that the docking subcomplex may be the true translocon, at least for these proteins, if not for other matrix cargoes as well. The RING subcomplex proteins are required for the export/release of receptors on the peroxisome membrane (Leon et al., 2006; Zhang et al., 2006). The dislocation/recycling of the receptors from the peroxisomes to the cytosol requires the action of a receptor recycling machinery, comprised of an E2-like ubiquitin-conjugating enzyme, Pex4p, two AAA ATPases, Pex1p and Pex6p, that interact with each other in an ATP-dependent manner, and a peroxisomal membrane protein (Pex15p in S. cerevisiae or PEX26 in mammals), which provides a docking site for Pex6p (Leon et al., 2006; Miyata and Fujiki, 2005; Platta et al., 2005). When this receptor recycling machinery is affected, a peroxisomal RADAR (acronym for Receptor Accumulation and Degradation in the Absence of Recycling) pathway becomes evident (Collins et al., 2000; Kiel et al., 2005a; Koller et al., 1999; Leon et al., 2006; Platta et al., 2004). This involves polyubiquitylation of peroxisome-membrane-associated Pex5p and Pex20p, most likely by redundant UBCs (Ubc1/Ubc4p/Ubc5p in S. cerevisiae), followed by their degradation by proteasomes (Kiel et al., 2005a; Kiel et al., 2005b; Kragt et al., 2005; Leon et al., 2006; Platta et al., 2004).
2. Shuttling receptors in peroxisomal matrix protein import.
a. Evidence for the shuttling of the PTS1 receptor, Pex5p
The identification and initial characterization of human PEX5 led to the discovery of a dual localization for this protein in mammalian cells (Dodt et al., 1995; Wiemer et al., 1995). Subcellular fractionation experiments indicated that PEX5 is predominantly present in the cytosolic fraction, but is also associated with the peroxisomes. This led to the hypothesis that PEX5 might be a mobile, cytosolic receptor that brings PTS1-containing cargo proteins to the peroxisome (Rachubinski and Subramani, 1995), by analogy to the SRP targeting cycle at the ER membrane (Pool, 2005; Walter and Blobel, 1981). In addition, a shuttling model of peroxisomal receptors had also been suggested for the yeast PTS2 receptor, Pex7p (see below, Marzioch et al., 1994).
A more careful characterization of the subcellular location of mammalian PEX5 showed that it exhibits a dynamic, as opposed to a static distribution, at the peroxisomal membrane: PEX5 localization at the peroxisome is affected by ATP availability and temperature in a reversible fashion (Dodt and Gould, 1996). In addition, using fibroblasts isolated from patients with peroxisome biogenesis disorders (PBDs), the authors observed that certain peroxins modulated this bimodal distribution: in particular, the RING peroxins, PEX2 and PEX12, appeared in this study as key determinants for the proper distribution of PEX5 between the peroxisome and the cytosol. These data suggested a dynamic cycling of PEX5 between the cytosol and the peroxisome.
However, no data clearly indicated whether PEX5 followed the simple shuttle model, where PEX5 delivers cargoes to the peroxisomal membrane prior to their translocation and never enters the matrix, or an extended shuttle where PEX5 enters into the peroxisomal lumen together with the cargo, unloads the cargo in the lumen and then recycles back to the cytosol (Kunau, 2001). Evidence that PEX5 follows the extended shuttle model came from work in mammalian cells (Dammai and Subramani, 2001).
This study also proved that modified PEX5 enters deep enough in the matrix to be cleaved by the intraperoxisomal protease. But how far PEX5 really enters the lumen is still a matter of debate (Erdmann and Schliebs, 2005; Kunau, 2001). PEX5 can temporarily behave as a membrane protein during the import cycle (Gouveia et al., 2003b; Gouveia et al., 2000), which could be consistent with either the extended shuttle model or the alternative “transient pore” model where PEX5 never leaves the membrane, but instead diffuses in the lipid bilayer from the docking site to the recycling site (Azevedo et al., 2004; Erdmann and Schliebs, 2005). According to this hypothesis, the peroxisomal translocon could be PEX5 itself or be comprised of PEX5 associated with PEX14. This hypothesis is suggested by the observation that S. cerevisiae Pex5p can spontaneously insert into a phospholipid bilayer (Kerssen et al., 2006), but this has only been shown to occur in vitro and the relevance of the insertion of Pex5p into an artificial bilayer to the physiological situation in vivo is unclear at present. In vitro import experiments using PEX5 and rat liver peroxisomes, followed by protease treatment to visualize the amount of imported receptor, defined several PEX5 populations engaged at different stages of the peroxisomal import cycle (Gouveia et al., 2003a). This includes PEX5 molecules in the stages 0 and 1 where PEX5 is protease-sensitive but located in the cytosol and on peroxisomes, respectively, “stage 2” peroxisome membrane-associated PEX5 molecules that are rendered 2-kDa shorter (at their N-termini) than the regular PEX5 protein upon protease treatment, and “stage 3” PEX5 molecules that are peroxisome-associated and fully protected from external protease. The fully-protected state of PEX5 (stage 3) was readily seen under ATP-limiting conditions, which we now know would likely allow receptor import into peroxisomes, but not export. These data suggest that PEX5 begins the import cycle in the cytosol (stage 0), then docks with membrane peroxins (stage 1), translocates into peroxisomes without requiring ATP (stage 3), and is then exported partially while persisting in the peroxisome membrane (stage 2), before it can be recycled back to the cytosol (stage 0) in an ATP-dependent manner (Gouveia et al., 2003a). The protease protection of a population of mammalian PEX5 during the import cycle in wild-type cells, as well as in mutants deficient in PEX5 recycling from the peroxisomes to the cytosol, has also been reproduced in an in vitro system that is capable of matrix protein import as well as PEX5 shuttling into and out of peroxisomes (Miyata and Fujiki, 2005).
b. Evidence for the shuttling of the PTS2 receptor, Pex7p
The concept of shuttling receptor first appeared after the observation that a fraction of the S. cerevisiae PTS2 receptor, Pex7p, associates with peroxisomes when bound to its cargo protein (Marzioch et al., 1994). Other results indicating that Pex7p may be an intra-peroxisomal receptor (Zhang and Lazarow, 1995; Zhang and Lazarow, 1996) were in fact caused by the tag that the protein carried (Nair et al., 2004). In S. cerevisiae (Rehling et al., 1996), P. pastoris (Elgersma et al., 1998), and mammals (Braverman et al., 1997; Mukai et al., 2002), Pex7p was found to be cytosolic and partially peroxisomal and therefore proposed to cycle in and out of peroxisomes. Definitive evidence of Pex7p cycling came from an elegant study in yeast (Nair et al., 2004). It is likely that the shuttling of Pex7p also applies in other organisms.
c. Evidence for the shuttling of the PTS2 auxiliary proteins, Pex18p/Pex20p
In lower eukaryotes, the PTS2 import pathway requires both Pex7p and a co-receptor or “PTS2 auxiliary protein”, Pex20p, the exception to the rule being S. cerevisiae, which possesses instead two redundant auxiliary proteins (Pex18p and Pex21p) (Purdue et al., 1998). Pex20p-like proteins interact with Pex7p and members of the docking complex, and some also interact with PTS2 sequences/cargo(es) (Otzen et al., 2005; Titorenko et al., 1998). In higher eukaryotes, the PTS2 auxiliary protein is substituted by a longer isoform of the PTS1 receptor, PEX5L, that contains an additional exon encoding a PEX7-binding domain (Dodt et al., 2001; Einwächter et al., 2001; Matsumura et al., 2000; Mukai et al., 2002; Otera et al., 2000). This feature is conserved in plants, where only the long isoform of Pex5p exists (Hayashi et al., 2005; Woodward and Bartel, 2005). Although the molecular details are unknown, PEX5L, like Pex20p-like proteins (Leon et al., 2006; Stein et al., 2002), is involved in the translocation of cargo-loaded PEX7 (Otera et al., 2000).
In addition to a common function, PTS2 auxiliary proteins are evolutionarily related to Pex5p in many ways. In particular, they share structural similarities, with a common Pex7p-binding motif (in the case of higher eukaryote’s PEX5L), common “docking motifs” made of diaromatic pentapeptide repeats (Wxxx[F/Y]), and a common N-terminal domain of about 30 residues (Dodt et al., 2001; Einwächter et al., 2001; Leon et al., 2006).
As expected for proteins with such striking similarities, several lines of evidence indicate that PTS2 auxiliary proteins also shuttle between the cytosol and the peroxisome during the import cycle. First, P. pastoris and Yarrowia lipolytica Pex20p and S. cerevisiae Pex18p display a dual subcellular localization, as does Pex5p (Leon et al., 2006; Purdue et al., 1998; Titorenko et al., 1998). Second, Pex20p and Pex5p share similar regulation and dynamics during the import cycle, as demonstrated in P. pastoris (Leon et al., 2006). Defects in the late steps of protein import, which lead to a failure in Pex5p recycling, also affect Pex20p localization and stability. Indeed, accumulation of receptors at the peroxisomal membrane triggers the peroxisomal RADAR pathway (see below), for which both Pex5p and Pex20p are the targets (Kiel et al., 2005a; Kiel et al., 2005b; Kragt et al., 2005; Leon et al., 2006; Platta et al., 2004; Platta et al., 2005). When its degradation is further prevented (e.g. by mutation of residue Lys19), P. pastoris Pex20p is mostly peroxisomal. In addition, Pex20p accumulates inside peroxisomes in mutants of the RING subcomplex (Leon et al., 2006, and unpublished results).
Contrasting results have been obtained in S. cerevisiae where it is not yet clear whether Pex18p and Pex21p enter peroxisomes during the import cycle (Purdue et al., 1998). Pex18p is constitutively degraded by the ubiquitin/proteasome pathway during the import cycle (Purdue and Lazarow, 2001). Interestingly, this constitutive degradation of Pex18p is abolished in several pex mutants in which the import of matrix proteins is blocked. This includes mutants lacking peroxins involved in receptor docking at the membrane (pex13∆ or pex14∆), a mutant that lacks the peroxisomal member of the E2 family of ubiquitin-conjugating enzymes (pex4∆), and others involved in receptor recycling (pex1∆) (Purdue and Lazarow, 2001). A reasonable possibility is that Pex18p is somehow deficient in recycling back to the cytosol and that it may be cleared from the peroxisome by the RADAR pathway (see section below). Indirect evidence in favor of the shuttling of Pex18p has been obtained: a chimeric protein made of its N-terminal half fused to the PTS1-binding domain (TPR repeats) of Pex5p could functionally substitute for Pex5p (Schäfer et al., 2004) in supporting PTS1 import. This indicates that the shuttling mechanism of Pex5p, if essential for its function as expected, is likely to be conserved for auxiliary proteins. However, it is not known whether the instability of Pex18p, on which its function may rely, was also transferred to the chimeric protein.
3. Steps in the import cycle of peroxisomal matrix proteins.
i. Cargo binding – Cargoes containing the PTS1 and/or PTS2 are synthesized in the cytosol, where they are bound by cytosolic receptors (with or without co-receptors or accessory proteins). The PTS1 sequence is bound directly by Pex5p, whereas the PTS2 sequence is bound by Pex7p, with Pex18p/Pex21p or Pex20p serving as co-receptors that might stabilize the receptor cargo complex (Leon et al., 2006; Stein et al., 2002). Pex20p has been reported to bind a synthetic peptide containing the PTS2 sequence from amine oxidase of H. polymorpha, and mutations in the PTS2-like sequence of Pex8p of P. pastoris abolish its interaction with PpPex20p (Zhang et al., 2006). Additionally, the thiolase of Y. lipolytica interacts with Pex20p using a region outside the PTS2 sequence (Smith and Rachubinski, 2001), so it is appropriate to treat Pex20p as a member of the cargo receptor family, despite the fact that it is not homologous to Pex7p.
ii. Docking – Following cargo binding, the receptors interact on the peroxisome membrane with the docking subcomplex. This is believed to be the first site of interaction. The docking of Pex5p and Pex20p with peroxisomes uses a common motif on these proteins, comprised of Wxxx(F/Y) repeats, for interaction with Pex14p on the peroxisome membrane. Pex5p interacts with both Pex13p (Bottger et al., 2000) and Pex14p (Albertini et al., 1997; Niederhoff et al., 2005) and so does Pex20p (Leon et al., 2006). These proteins only interact indirectly with Pex17p (Leon et al., 2006). Pex5p and Pex20p also interact with Pex8p, which is an intraperoxisomal peroxin. Pex7p can also interact with Pex13p (Stein et al., 2002) and Pex14p (Niederhoff et al., 2005; Stein et al., 2002), independently of Pex18p/Pex21p (Stein et al., 2002) but no interaction has been reported between Pex7p and Pex8p.
iii. Translocation – As stated earlier, the membrane translocation step for PTS receptors is operationally defined as a peroxisome-associated and protease-resistant state, which could mean either that the receptor enters the peroxisome lumen or is deeply embedded in the peroxisome membrane, from where it can release cargo into the peroxisome matrix. At steady-state, a pool of Pex5p, Pex7p and Pex20p is peroxisome-associated and protease protected. Experiments performed in vivo suggest that the peroxisomal association of these receptors requires the presence of the central docking subcomplex component, Pex14p. In its absence, Pex20p is exclusively cytosolic (Leon et al., 2006), Pex5p is not peroxisome associated (Zhang et al., 2006) and Pex7p shows reduced binding to peroxisomes (Girzalsky et al., 1999). Therefore, translocation of the receptors into or across the peroxisomal membrane requires Pex14p, and probably the docking subcomplex.
iv. Cargo release – Not much is known about cargo release from the receptor-cargo complexes. In vitro experiments with HpPex8p suggest that it can release cargo from HpPex5p (Wang et al., 2003). This has led to the idea that the intraperoxisomal Pex8p, which interacts with Pex5p and Pex20p, may be involved in cargo release. An attractive model for such cargo displacement is the presence of PTS2 and/or PTS1 sequences on Pex8p. Unfortunately, mutation of each of these PTSs on Pex8p does not affect the function of Pex8p (Waterham et al., 1994) and not all organisms containing Pex8p have both PTSs (Zhang et al., 2006), making this simple model untenable. Furthermore, this protein is found only in fungi and direct evidence for the involvement of Pex8p in cargo release is also lacking for the PTS2 pathway.
v. Retrotranslocation – The extended shuttle model for PTS receptor dynamics suggested that these receptors might have cis-acting export sequences and also require trans-acting factors for their exit from peroxisomes to the cytosol (Dammai and Subramani, 2001). Both of these predictions appear to be true for Pex5p and Pex20p. For example, human PEX5 lacking its first 17 amino acids was capable of binding cargo and translocating to peroxisomes, but was deficient in the ATP-dependent export of PEX5 from peroxisomes to the cytosol, in an in vitro system (Costa-Rodrigues et al., 2004). Similarly, the first 19 amino acids of PpPex20p are required for its recycling from peroxisomes to the cytosol (Leon et al., 2006).
The translocated states of Pex5p and Pex20p are characterized by being protease-protected and inaccessible to the cytosolic machinery required for the RADAR pathway, meaning that these receptors are not available for polyubiquitylation and subsequent proteasomal degradation. Following this state, the receptors become accessible to the recycling and RADAR machineries (Leon et al., 2006). We refer to the transition from the protease-protected (intraperoxisomal) state to the RADAR and recycling machinery-accessible state as retrotranslocation or export. The details of this step are poorly understood. However, for PpPex20p, we know that the RING subcomplex components, Pex2p, Pex10p and Pex12p, are each necessary for this export (Leon et al., 2006). In P. pastoris cells lacking any of these components, the other two components are unstable and the RING subcomplex is not assembled efficiently (Hazra et al., 2002; Zhang et al., 2006). Furthermore, in each of these mutants, PpPex20p-GFP is peroxisome-associated and also stable (i.e. inaccessible to the RADAR machinery) (Leon et al., 2006). In the absence Pex2p of P. pastoris, Pex5p and Pex8p are also peroxisome-associated and protease-protected, suggesting that the RING peroxins are not required for the import of these proteins into peroxisomes (Zhang et al., 2006), and consistent with the notion that accumulation of the receptors in peroxisomes is the result of a block in their export. These results suggest that the RING subcomplex is required for export of Pex5p and Pex20p from peroxisomes, but whether it serves as the retrotranslocon, or indirectly modulates the function of the same translocon used for receptor import is unclear.
vi. Recycling or dislocation from the peroxisome membrane – Several reports point to the requirement of the peroxins, Pex4p, Pex22p, Pex1p and Pex6p for the recycling or dislocation of Pex5p from peroxisomes to the cytosol in yeast (Collins et al., 2000; Miyata and Fujiki, 2005; Platta et al., 2005; van der Klei et al., 1998). The same trans-acting factors required by Pex5p also play a role in Pex20p recycling (Leon et al., 2006). The use of in vitro systems has allowed the reconstitution of the dislocation step from peroxisomes to the cytosol for yeast and mammalian Pex5p (Costa-Rodrigues et al., 2004; Miyata and Fujiki, 2005; Platta et al., 2005). These studies show a requirement for ATP hydrolysis, most likely by the AAA ATPases, Pex1p and Pex6p, for this step (Platta et al., 2005).
It should be noted that the peroxisome membrane of fungi contains several components (putative RING E3-like ligases, an E2-enzyme Pex4p, and two AAA-family ATPases) with striking parallels to the ERAD (ER-associated degradation) pathway that requires an E3 ligase (Hrd1p), an E2-like enzyme (Ubc7p) and a AAA-family ATPase (Cdc48p) (Romisch, 2005). In both systems, the E3 ligase (still putative for peroxisomes) is an integral-membrane protein of the organelle, the E2 is anchored at the appropriate membrane by association with integral membrane proteins (Cue1p for ERAD and Pex22p for peroxisomes), and the AAA ATPase is localized to the organelle by interaction with membrane-associated proteins (Ubx2p, Der1p for ERAD and Pex15p or PEX26 for peroxisomes). However, while ERAD is a degradative pathway used to destroy aberrant or unwanted proteins, peroxisomal receptor recycling presumably serves to protect the receptors from the proteasome, so that they can catalyze additional rounds of cargo import.
vii. RADAR – When recycling of the PTS receptors is blocked by mutations in the receptor recycling machinery, then receptors would accumulate on the peroxisome membrane and could block upstream events including cargo and receptor import into peroxisomes. Results from several laboratories reveal the existence of a pathway related to a quality-control system that clears the peroxisome membrane of receptors that cannot be recycled after a round of import. By analogy to the acronym “ERAD”, we have termed this peroxisome-related machinery the “RADAR” pathway, standing for Receptor Accumulation and Degradation in Absence of Recycling, which is also clearer than “quality-control pathway” that is often used for handling of misfolded proteins. This pathway is present in all organisms, from yeasts to plants and mammals (Collins et al., 2000; Kiel et al., 2005a; Platta et al., 2004). The RADAR pathway, which has been studied only for Pex5p and Pex20p, appears to involve similar mechanisms for both proteins. A lysine near the N-termini of these proteins (K21 in HpPex5p, K22 in PpPex5p and K19 in PpPex20p) appears to be the target for polyubiquitylation, not by Pex4p, but rather by some other Ubc (e.g. Ubc1p, Ubc4p and/or Ubc5p in S. cerevisiae) (Kiel et al., 2005a; Kiel et al., 2005b; Kragt et al., 2005; Leon et al., 2006; Platta et al., 2004). This polyubiquitylation involves a K48-linkage between the ubiquitin moieties. Following polyubiquitylation, these proteins are degraded by the proteasome, because blocking proteasomal activity with inhibitors such as MG132, stabilizes these proteins (Kiel et al., 2005b). The robustness of this RADAR pathway may vary between organisms (Collins et al., 2000; Kiel et al., 2005a; Platta et al., 2004). This is illustrated by the fact that in P. pastoris, plants and mammals, PEX5 is completely degraded by RADAR when recycling is blocked, whereas in S. cerevisiae a significant amount of Pex5p remains even when recycling is compromised, but instead appears to be strongly polyubiquitylated (Collins et al., 2000; Kiel et al., 2005a; Leon et al., 2006; Platta et al., 2005). This provides the long-sought explanation for the instability of PEX5 in human patient cell lines (Yahraus et al., 1996).
References |
PEROXISOME TURNOVER BY MICROPEXOPHAGY
– AN AUTOPHAGY-RELATED PROCESS
Many organisms stringently regulate the number, volume and enzymatic content of peroxisomes (and other organelles). Understanding this regulation requires knowledge of how organelles are assembled and selectively destroyed, in response to metabolic cues. In the past decade, considerable progress has been achieved in the elucidation of the roles of genes involved in peroxisome biogenesis, half of which are affected in human peroxisomal disorders. The recent discovery of intermediates and genes in peroxisome turnover by selective autophagy-related processes (pexophagy), opens the door to understanding peroxisome turnover and homeostasis.
During autophagy, cargoes comprised of individual proteins, bulk cytosol and/or subcellular organelles, are degraded in the lysosome (or vacuole in yeast), with the ensuing recycling of the amino acid and lipid precursors. Although autophagy was initially thought to enable cells to survive nutrient starvation by degradation and recycling of dispensable cellular components, it is now recognized to be involved in a plethora of cellular responses including the regulation of organelle number (Tolkovsky et al., 2002; Tuttle and Dunn, 1995; Yokota, 2003), development (Levine and Klionsky, 2004; Melendez et al., 2003; Otto et al., 2003), cell death (Hernandez et al., 2003), lifespan (Melendez et al., 2003), bacterial pathogenesis (Hernandez et al., 2003), and cancer (Ogier-Denis and Codogno, 2003). Not surprisingly, its involvement in pathological states and disease progression in mammals is rapidly gaining attention and interest (Cuervo, 2004).
Autophagy is universal to all eukaryotic cells, including yeasts, worms, insects, plants and mammals (Klionsky, 2004). In unicellular organisms, such as yeasts, it is observed primarily under nutrient starvation conditions or during the re-adaptation of cells switched from certain environments to others. Two morphologically distinct, but mechanistically related, forms of autophagy common to uni- and multi-cellular eukaryotes have been described. These are macroautophagy and microautophagy (Fig. 1). A third form, called chaperone-mediated autophagy, has been found only in mammalian systems and is described elsewhere (Cuervo, 2004). Autophagy is often a degradative process, but it can be either selective or non-selective with respect to cargo that is turned over. The degradation of non-selective cargo is a mechanism for recycling any excess cellular components (e.g. cytosol and/or organelles) and may be a strategy for adaptation to different environments. In contrast, cells resort to selective autophagic turnover of non-functional or damaged organelles, or protein aggregates, that might be too large for other proteolysis machinery, such as the proteasome.
Unlike the autophagic pathways, the related cytosol-to-vacuole transport (Cvt) pathway, described to date only in Saccharomyces cerevisiae, delivers a limited set of specific, oligomeric, vacuolar hydrolases, aminopeptidase I (ApeI) and a–mannosidase 1 (Ams1) directly from the cytosol to the vacuole (Abeliovich and Klionsky, 2001; Stromhaug and Klionsky, 2001)(Fig. 1). This mode of vacuolar delivery may be used, rather than the usual route via the secretory pathway, by proteins that fold and oligomerize rapidly in the cytosol. Their folded/oligomerized state may be incompatible with translocation across the ER membrane, which transports only unfolded proteins.
The microautophagy, macroautophagy and Cvt pathways, while being morphologically distinct, share many proteins (Mukaiyama et al., 2002; Sakai et al., 1998; Stromhaug et al., 2001). However, each of these processes also uses unique proteins (Abeliovich and Klionsky, 2001; Mukaiyama et al., 2002), which likely account for the types of cargo selected by each process, the physiological signals activating these pathways, and morphological differences between them.
Pexophagy refers to the specific turnover of peroxisomes by autophagy. However, because pexophagy can occur by processes resembling either macroautophagy or microautophagy, these turnover mechanisms are referred to as macropexophagy (Kiel and Veenhuis, 2004) and micropexophagy (Habibzadegah-Tari and Dunn, 2004), respectively, to distinguish the distinct mechanisms and proteins involved.
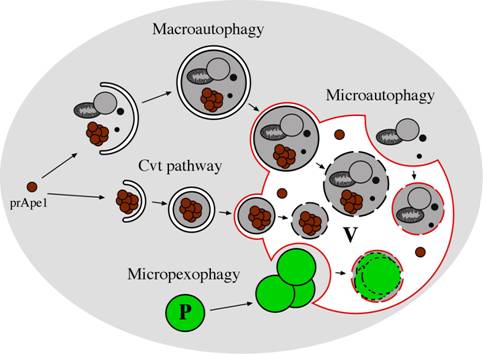
Figure 1: Morphological steps in the Cvt, macroautophagy and microautophagy pathways in yeast. In microautophagy, organelles and/or cytosolic proteins are engulfed by the vacuolar membrane in a Pac-Man-like fashion and degraded in the vacuole lumen. Macroautophagy involves the formation of large (300-900 nm) cytosolic, double-membrane vesicles (autophagosomes), which sequester organelles and/or cytosolic proteins. Once formed, autophagosomes fuse with the vacuole, releasing a single-membrane vesicle (autophagic body) into the lumen of this organelle where the autophagic bodies are degraded. In the Cvt pathway, which is biosynthetic rather than degradative, cytosolic cargo, such as the precursor of aminopeptidase I (prApe1), is first oligomerized in the cytosol and engulfed by a double-membrane to generate a Cvt vesicle (140-160 nm). These vesicles fuse with the vacuole, yielding intravacuolar Cvt bodies that are degraded, releasing prApe1 into the lumen, where it is processed to the mature form. Selective turnover of peroxisomes can occur by macroautophagy- or microautophagy-like processes called macropexophagy and micropexophagy, respectively. P: peroxisome, V: vacuole.
Recently a new nomenclature was adopted for autophagy-related (ATG) genes (Klionsky et al., 2003). To date 30 ATG genes have been identified and several genes involved in other cellular pathways (e.g. vacuolar protein sorting or VPS) also play a role in autophagy-related processes. Somewhat surprisingly, although micropexophagy appeared to be a morphologically-distinct process from the macroautophagy and Cvt pathways, it was found upon closer analysis to share many proteins with other autophagy-related processes. Additionally, there are also proteins uniquely involved in micropexophagy.
Pichia pastoris as a model for autophagy-related processes
Among the eukaryotes studied to date, P. pastoris is unique in having all of the autophagy-related processes (macroautophagy, micropexophagy, macropexophagy, the Cvt pathway and most likely microautophagy). Given the substantial overlap between these pathways, the use of this yeast allows us to elucidate the role of specfic ATG and other genes in each of these pathways. Additionally, the ability to induce peroxisomes on a variety of carbon sources, as well as the ability to induce either micropexophagy or macrophagy at will, permit us to study all forms of pexophagy in a single organism. Finally, the availability of pexophagy mutants and the genome of P. pastoris provide rapid access to the genes and proteins required for pexophagy. These studies have revealed the involvement in pexophagy of Ser/Thr kinases, PI-3 kinase, PI-4 kinase, ubiquitin-like molecules and their conjugating systems, sterol glucosyltransferases, and selectivity factors for pexophagy.
Micropexophagy
Micropexophagy was monitored by fluorescence and electron microscopy, following a shift of P. pastoris cells from methanol to glucose (Sakai et al., 1998; Tuttle and Dunn, 1995). The vacuole was labelled with the vital dye, FM4-64, which is taken up by endocytosis. Peroxisomes were tagged by the transport of GFP, fused at its C-terminus to a peroxisomal matrix targeting signal, termed PTS1. From an analysis of the events and kinetic intermediates observed, a model was proposed for micropexophagy (Fig. 2A) and non-conditional mutants blocked at various steps were identified (Fig. 2B) (Mukaiyama et al., 2002; Sakai et al., 1998; Stromhaug et al., 2001). The peroxisomes and a single spherical vacuole are juxtaposed next to each other initially (Stage 0). Upon induction of micropexophagy, the vacuole invaginates (Stages 1a-1c), and extends two arms around the peroxisome. During Stage 1 the vacuole usually septates to generate multiple vacuolar compartments, which then coordinately engulf a cluster of peroxisomes. The septated vacuoles or the extended vacuolar arms then fuse around the peroxisomes to engulf it completely (Stage 2), followed by the vacuolar degradation of the peroxisomal membrane and its contents (Stage 3). Previous studies have shown that during micropexophagy in P. pastoris, mitochondria are not degraded, but cytosolic formate dehydrogenase was turned over (Tuttle and Dunn, 1995).
The dissection of the morphological steps in micropexophagy and the availability of many mutants in the process allowed the isolation and characterization of the genes involved and the determination of where particular gene products act during micropexophagy (Mukaiyama et al., 2002; Stromhaug et al., 2001). Examination of the genes affecting micropexophagy reveals two themes – (a) many are also needed for other autophagy-related processes, such as the Cvt and macroautophagy pathways, and (b) some genes are unique to micropexophagy, and dispensable for related pathways. The determination of the steps in micropexophagy and the identification of the proteins has facilitated a detailed molecular analysis of the functions of the proteins involved.
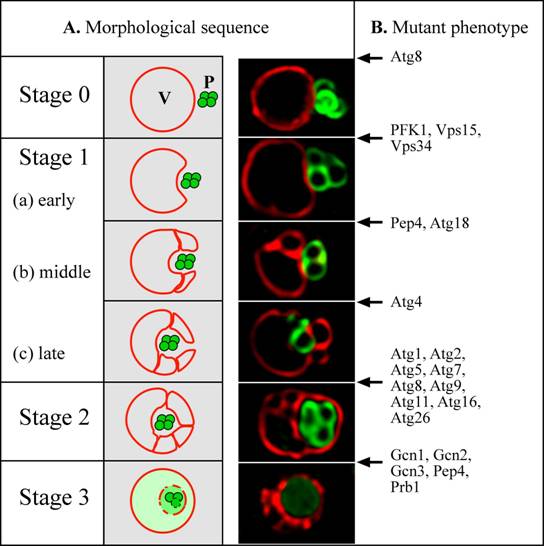
Figure 2: Model for micropexophagy in P. pastoris (Mukaiyama et al., 2002; Sakai et al., 1998). (A) Morphological intermediates and stages. Stage 0 – Peroxisomes and vacuoles are juxtaposed. Stage 1 - peroxisomes start to become engulfed by invagination of the vacuole membrane. This process proceeds through (a) early, (b) middle and (c) late stages. Stage 2 - homotypic fusion of the vacuole membrane causes the peroxisomes to become completely surrounded within a subvacuolar vesicle. Stage 3 - the subvacuolar vesicle and peroxisomes start to degrade, and are completely broken down in the vacuole lumen. A simple invagination of the vacuolar membrane appears to be sufficient to engulf a small peroxisomal cluster, but large peroxisomal clusters are engulfed by multiple, septated vacuoles (Mukaiyama et al., 2002). In the fluorescence picture, peroxisomes were tagged with the GFP-SKL fusion (Green) and the vacuole was labeled with FM4-64 (Red). (B) Arrest points of micropexophagy mutants in P. pastoris (Guan et al., 2001; Kim et al., 2001; Mukaiyama et al., 2002; Sakai et al., 1998; Stromhaug et al., 2001; Yuan et al., 1999; Yuan et al., 1997) (our unpublished data). A few proteins with dual arrest points (e. g. Atg8 and Pep4) are likely to have dual functions (see text). P: peroxisome, V: vacuole.
Role of a PAS-like membrane compartment in micropexophagy
It was anticipated that the genes involved in the early steps of micropexophagy and the macroautophagy/Cvt pathways, would be distinct, whereas those acting at the late vacuolar hydrolysis steps would be common. Surprisingly, however, components of the NPS (e.g. Atg9), and many proteins needed for the formation of the PAS, were also needed for micropexophagy (Mukaiyama et al., 2002; Stromhaug et al., 2001). To explain this anomaly, it was speculated that a PAS-like structure might be necessary for the terminal fusion of the vacuolar arms around the cargo (Subramani, 2001). Experimental evidence for the involvement of such a structure, called micropexophagy apparatus (MIPA) was published recently and this structure has been proposed to be required for complete peroxisome engulfment (Mukaiyama et al., 2004). The MIPA contains proteins (Atg8 and Atg26) needed for both micropexophagy and macropexophagy (Oku et al., 2003). The location of the MIPA is also interesting in that it is positioned on the side of the peroxisome furthest away from the site of initial juxtaposition of the peroxisome and the vacuole (Fig. 4). If the MIPA and PAS are indeed related structures, then it is easy to understand why many of the proteins required for the formation of the PAS are also needed for micropexophagy.
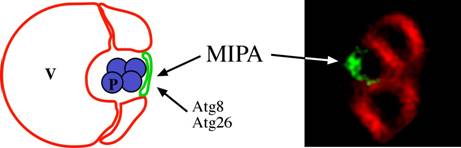
Figure 3: Localization of the MIPA during micropexophagy. In the fluorescence picture, the MIPA was localized with GFP-PpAtg8 fusion (Green) and the vacuole with FM4-64 (Red) (Mukaiyama et al., 2004). The only two proteins currently localized to the MIPA in P. pastoris are Atg8 and Atg26 (Mukaiyama et al., 2004; Oku et al., 2003). V: vacuole, P: peroxisome
Areas of ongoing research in pexophagy
Autophagy-related processes in P. pastoris
Mechanism of pexophagy in P. pastoris
Selectivity of pexophagy
Role of phosphatidylinositol kinases, phosphoinositide lipids and lipid-binding proteins in pexophagy
Link between peroxisome turnover and biogenesis
References
SELECTIVE AUTOPHAGY OF PEROXISOMES (PEXOPHAGY) IN MAMMALIAN CELLS
While yeast cells have greatly contributed as model systems to decipher the mechanisms and molecular players underlying selective autophagy pathways (such as pexophagy), the molecular basis for regulation and specificity of these processes is poorly understood in mammalian cells. The pivotal role of autophagy pathways for cellular homeostasis under physiological, as well as under various stress conditions, is emphasized by the well documented contribution of autophagy to various human diseases, including inflammation, cancer, metabolic disorders, neurodegeneration and aging (Klionsky, 2010; Levine et al., 2011; Rabinowitz and White, 2010). In order to understand and ultimately manipulate this highly effective degradation machinery, it is crucial to broadly assess the functional architecture of the involved gene networks of the associated processes. In addition, an assessment of the evolutionary conservation of the relevant sub-processes and functional modules will help to evaluate the utility of model systems, such as yeast, for in-depth molecular studies. In this subproject we are therefore aiming to understand on a mechanistic level the processes regulating selective degradation of cytosolic cargoes (e.g. damaged or superfluous organelles such as peroxisomes, misfolded proteins, intracellular pathogens) by the autophagy/lysosome system in mammalian cells. Using bioinformatical data acquisition and gene network analyses, we have established a human extended autophagy network which comprises roughly 1,200 human genes and over 5,000 interactions.
Subcellular organization of the AXAN. The prime cellular compartment for each gene was determined based on GO terminology and assigned as a node attribute. The network was displayed using attribute grouping in Cytoscape. CYS: cytosol, CSK: cytoskeleton, ENDO: endosome, EXTRA: extracellular, GA: Golgi apparatus, LYSO: lysosome, MITO: mitochondrion, NUCL: nucleus, PEX: peroxisome, PM: plasma membrane, RIB: ribosome, N/A: not applicable. The unconnected nodes have not been integrated into the layout. Interactions: green; protein-protein interactions, light green; MiMI interaction data, red; transcriptional regulation data).
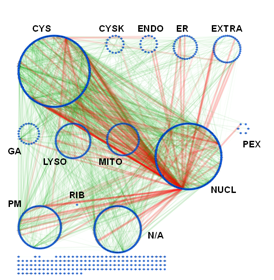
This data set is used to set up and refine hypothesis-driven experimental studies on selected aspects of selective autophagy pathways. In particular, we hope to address evolutionary conservation and functional network architecture of the entire gene network, as well as the role of selectivity factors and intracellular signaling pathways for the selective degradation of cytosolic targets.
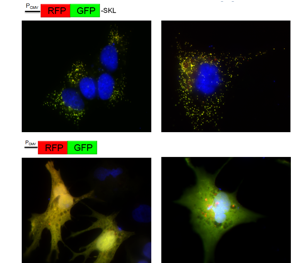
Human NT2 cell line transfected to overexpress fusion proteins RFP-GFP-SKL (localizing to peroxisomes, top panel) or RFP-GFP (cytosolic, bottom panel). Different treatments (e.g. starvation in EBSS buffer, right panels) may induce changes in the relative contribution of intact and fragmented reporter protein due to autophago/lysosomal degradation (as indicated by accumulation of red RFP fragment).
This project is funded through a seed grant (to Andreas Till and Suresh Subramani) by The San Diego Center for Systems Biology (SDCSB), one of the NIGMS-funded National Centers for Systems Biology.
http://sdcsb.org
http://www.systemscenters.org/
Klionsky, D.J. (2010). The autophagy connection. Dev Cell. 19:11-2.
Levine, B., N. Mizushima, and H.W. Virgin. (2011). Autophagy in immunity and inflammation. Nature. 469:323-35.
Rabinowitz, J.D., and E. White. (2010). Autophagy and metabolism. Science. 330:1344-8.
|


